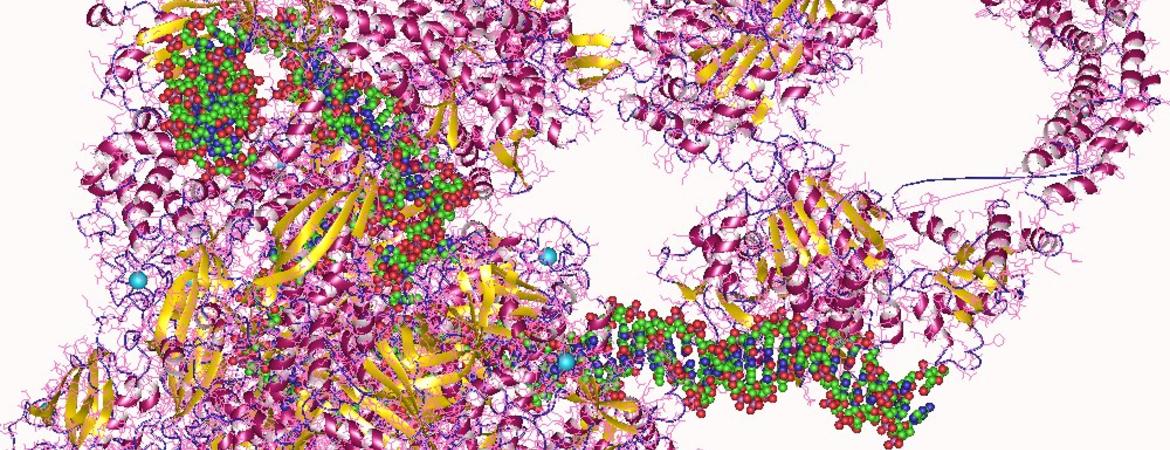
Gregor Blaha, an assistant professor of biochemistry at UC Riverside, is a coauthor on a research paperpublished last week in the journal Science that reports on the cryo-electron microscopy structures of several complexes of RNA polymerase bound to the ribosome.
In all living cells, RNA polymerase transcribes DNA into RNA and the ribosome translates the RNA into proteins. RNA polymerase and the ribosome are two of the most well-studied proteins in all of molecular biology, but almost all studies on these proteins look at them individually instead of them working together as they do in the cell. How these proteins interact with one another has puzzled scientists for decades.
"Our work is a breakthrough in the emerging field of transcription-translation coupling, a field that promises to provide a better understanding of gene expression in bacteria,” said Blaha, who provided key material for this study and was involved in the interpretation and discussion of the results. “It opens up new venues to target antibiotic-resistant bacteria.”
To image these complexes, the researchers first isolated all components of these complexes individually, before bringing them together into defined complexes and preparing samples for imaging in a cryo-electron microscope.
Blaha explained that in the imaging process, millions of individual images are collected and then combined into three dimensional representations before the structures of these complexes can be built.
“Then the fun part begins, the analysis of these structures and figuring out what they are telling us,” he said.
The structures show for the first time how transcription and translation may be coordinated on one RNA. According to Blaha, this is significant because polymerase extends the RNA one nucleotide at a time, while the ribosome moves three nucleotides every time it extends the protein it synthesizes by one amino acid. Inevitably both processes need to be coordinated, he said, to allow successful translation of the genetic information from DNA into proteins.
“Although the coupling of transcription and translation has been known since the dawn of molecular biology, only now are the tools available to actually elucidate what brings about the coordination of both,” he said. “This has generated much interest in the community which can best be seen in the increase of publications in the last years — three publications in Science last month alone.”
Now that Blaha and his colleagues have an idea of how transcription and translation are coordinated, they are interested in figuring out how the coordination is actually set up.
Blaha received his doctoral degree in 2001 from the Vienna University of Technology in Austria, following which he was a postdoctoral researcher at Yale University. An expert on the interplay between protein translation and transcription, he joined UCR in 2012.
Besides Blaha, the research paper’s coauthors are scientists at Rutgers University in New Jersey and the University of Michigan at Ann Arbor headed by Richard Ebright and Min Su, respectively. Blaha was supported by University of California discretionary funds.
The title of the research paper is “Structural basis of transcription-translational coupling.”
____________________________________
Thumbnail image: A depiction of a human RNA polymerase from Wikimedia Commons.
Read the original article here:
